Why we need Markov Models
Consider a scenario where we have four students who get together in pairs to work on the homework for a class.
1. Alice and Bob
2. Bob and Charles
3. Charles and Debbie
4. Debbie and Alice.
Alice and Charles just can’t stand each other, and Bob and Debbie had a relationship that ended badly.
In this example, the professor accidentally misspoke in class, giving rise to a possible misconception among the students in the class. Each of the students in the class may subsequently have figured out the problem, perhaps by thinking about the issue or reading the textbook.
In subsequent study pairs, he or she may transmit this newfound understanding to his or her study partners.
We therefore have four binary random variables, representing whether the student has the misconception or not
Because Alice and Charles never speak to each other directly, we have that A and C are conditionally independent given B and D. Similarly, B and D are conditionally independent given A and C.
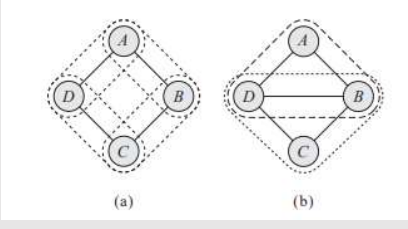
We can try to represent this distribution using Bayesian Network, but it encodes the independence assumption that (A ⊥ C | {B, D}). However, it also implies that B and D are independent given only A, but dependent given both A and C. Hence, it fails to provide a perfect map for our target distribution.
Another attempt, shown in figure (c), is equally unsuccessful. It also implies that (A⊥ C | {B, D}), but it also implies that B and D are marginally independent.
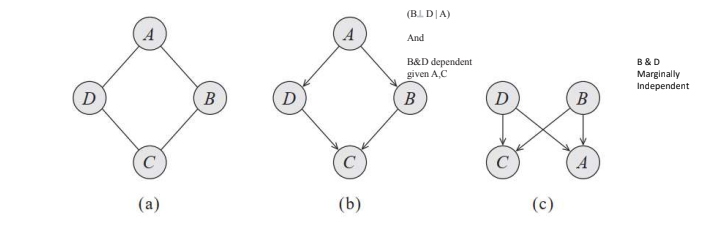
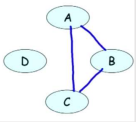
(a) Represents study pairs over 4 students
(b) Represents first attempt at bayesian network model
(c) Represents second attempt at bayesian network model
All other candidate BN structures are also flawed, so that this distribution does not have a perfect map.
Independencies (A ⊥ C | {B, D} and (B⊥ D | {A,C} cannot be naturally captured in a Bayesian network.
Bayesian network requires that we ascribe a directionality to each influence.
In this case, the interactions between the variables seem symmetrical, and we would like a model that allows us to represent these correlations without forcing a specific direction to the influence.
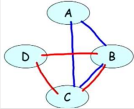
Markov Network: A representation that implements this intuition is an undirected graph. As in a Bayesian Network, the nodes in the graph of a Markov network represent the variables, and the edges
correspond to a notion of direct probabilistic interaction between the neighboring variables (an interaction that is not mediated by any other variable in the network).
In this case, the graph of figure (a), which captures the interacting pairs, is precisely the Markov network
structure that captures our intuitions for this example.
The remaining question is how to parameterize this undirected graph.
Because the interaction is not directed, there is no reason to use a standard CPD, where we represent the distribution over one node given others. Rather, we need a more symmetric parameterization.
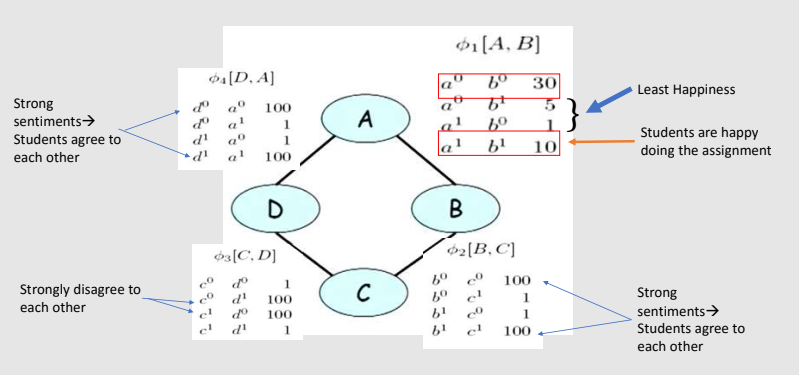
Intuitively, what we want to capture is the affinities between related variables.
For example, we might want to represent the fact that Alice and Bob are more likely to agree than to disagree.
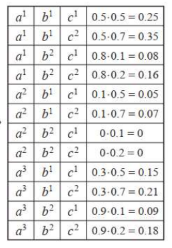
We associate with A,B a general-purpose function, also called a factor:
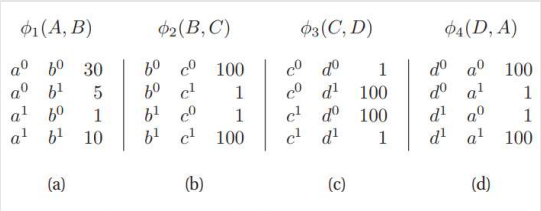
Hence we introduce the Φ function. It is also known as the affinity function or the compatibility function or the soft constraint
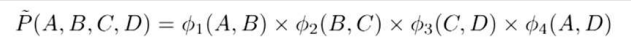
But this formula as normal probability won’t work here as the values are not in binary or continuous values. So, it's an unnormalized measure. Hence the tilde ~ on P.
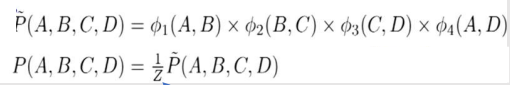
Z is called Partition Function or normalizing constant.
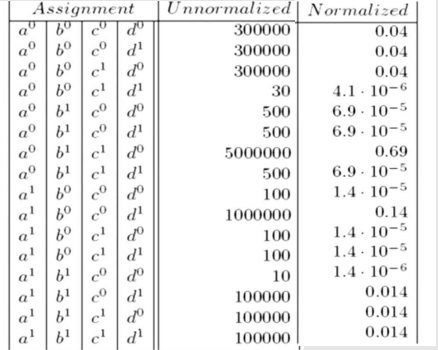
If we divide Unnormalized values by Z then we get Normalized Values.
Z = sum of all un-normalized values
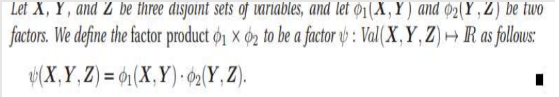
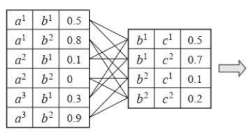
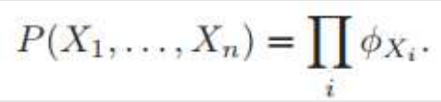
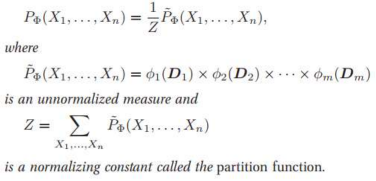
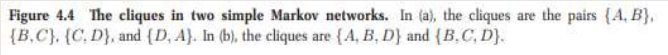
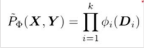
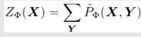
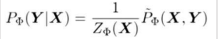
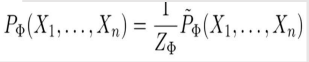
 This is called Family of Conditional distribution over X.
This is called Family of Conditional distribution over X.